 Gene Set Analysis d'enrichissement est l'une des nombreuses approches de la analyse de l'expression du gène les données de profil et est décrit dans un papierdes travailleurs du Broad Institute.
Gene Set Analysis d'enrichissement est l'une des nombreuses approches de la analyse de l'expression du gène les données de profil et est décrit dans un papierdes travailleurs du Broad Institute.
Le concept de base a été motivée par le constat que l'étude gènes individuels montrant la différence la plus significative du niveau d'expression entre deux états ou des phénotypes est manque de vision mécaniste. Plutôt, il est plus logique de prendre une un ensemble de gènes le partage de certaines lien biologique, et poser la question - ne l'ensemble montré aucune statistique enrichissement significatif dans les gènes qui ont une expression différentielle?
A ensemble des gènes peut être choisi, a priori, pour un certain nombre de raisons e.g. l'ensemble des gènes connus pour être influencé par les cours- ou une sous-expression d'un micro-ARN, ou peut-être un ensemble choisi en fonction de l'emplacement chromosomique, ou des gènes pour lesquels la fonction moléculaire, composant cellulaire et / ou processus biologique ont été affectés à l'aide des vocabulaires contrôlés de l' Gene Ontology.
Un avantage de l'approche GSEA est qu'il est possible d'intégrer votre ensemble de données complètes, non seulement ces transcriptions avec un seuil d'expression différentielle choisie arbitrairement. Je suis sûr que beaucoup de gens qui lisent cet vont penser - «Comment peut-il être acceptable d'utiliser l'ensemble des données? Normalement, je n'envisagerait gènes avec >2 (Ou une autre valeur préférée)-expression différentielle fois. "La raison pour laquelle l'approche est valable, c'est que les gènes exprimés à des niveaux bas ou avec un grand écart entre les répétitions ne contribuent pas à la principale mesure utilisée par GSEA, l''Note d'enrichissement» (ES).
GSEA agit en premier classement la valeur de l'expression pour chaque gène par Signal to Noise Rapport - calcul de la différence entre les valeurs moyennes d'échantillons représentant chacun le phénotype et leur mise à l'échelle par la somme des écarts-types. Cela signifie que les gènes avec de grandes différences dans le niveau d'expression entre les différents états et peu de variation entre les répétitions biologiques sont classés hautement.
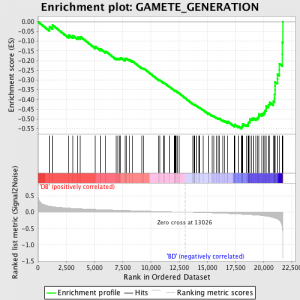
La prochaine étape est que l'ES, la statistique primaire générée par GSEA, est calculée pour chaque ensemble de gènes - dans le manuel GSEA, qui documente le logiciel excellemment, il déclare:
«Tous les gènes sont d'abord classés selon leur rapport signal sur bruit, puis l'ES est calculée en "marchant" en bas de la liste de classement des gènes croissant une rodage somme statistique quand un gène se trouve dans l'ensemble des gènes et réduction quand il n'est pas. Le ampleur de l'incrément dépend de la corrélation du gène avec un phénotype. L'ES est la déviation maximum à partir de zéro rencontrés dans la marche de la liste. A positif ES indique gène ensemble enrichissement à l' supérieur de la liste de classement; une négative ES indique gène ensemble enrichissement à l' inférieur de la liste de classement. "
Les valeurs de ES sont normalisée basé sur le gène taille de l'ensemble, puis une taux de fausses découvertes est calculé, de donner une estimation de la probabilité de faux positifs. GSEA utilise une valeur par défaut très détendue 25%, qui est apte à la génération d'hypothèse d'un nombre relativement important de réplique biologique.
Les scientifiques qui travaillent sur les données de non-human échantillons peuvent toujours utiliser GSEA, mais il fautméfiez-vous - L' symboles de gènes utilisé par GSEA sont "traduit»À partir de leur i.e des équivalents humains. identifiants utilisés pour les gènes de vos espèces d'intérêt représenté sur la puce sont converties en symboles pour leur orthologues humains, ensuite utilisée dans l'analyse. Subramanian et ses collègues réclamer que cette conversion a peu ou aucun effet sur l'utilité de GSEA; il a été utilisé avec succès chez de multiples espèces non-humaines, mais bien sûr, cela doit être gardé à l'esprit lorsqu'ils enquêtent sur les résultats en détail.
Pour une excellente, en profondeur, examen des outils de la voie, consulter:
Une autre bonne source de conseils sur l'analyse des voies, surtout pour ceux qui sont familiers avec le paquet statistiques R est ici.
Pour en savoir plus