 Gene Set Anrikning Analys är ett av många sätt att den analys av genuttryck profildata och beskrivs i ett papperfrån arbetare vid Broad Institute.
Gene Set Anrikning Analys är ett av många sätt att den analys av genuttryck profildata och beskrivs i ett papperfrån arbetare vid Broad Institute.
Grundtanken föranleddes av observationen att studera enskilda gener visar den mest betydande skillnaden i uttryck nivå mellan två stater eller fenotyper är saknar mekanistiska inblick. Istället, det är mer förnuftigt att ta en uppsättning av gener dela några biologisk länk, och ställa frågan - har hela uppsättningen visade någon statistiskt signifikant anrikning i de gener som har differentiellt uttryck?
ETT gen uppsättning kan väljas, a priori, för ett antal skäl e.g.. ut de kända gener som skall påverkas av över- eller under-uttryck av en mikro-RNA, eller kanske en uppsättning väljs utifrån kromosomalt läge, eller gener för vilka molekylära funktion, cellulär komponent och / eller biologisk process har tilldelats med hjälp av kontrollerade vokabulärer i Genontology.
En fördel till GSEA tillvägagångssätt är att det är möjligt att införliva din fullständig uppsättning uppgifter, inte bara de transkript med en godtyckligt vald differentiellt uttryck tröskel. Jag är säker på att många människor som läser detta kommer att tänka - "Hur kan det vara OK att använda den kompletta databas? Normalt skulle jag bara tänka gener med >2 (Eller annan favorit värde)-faldig differentiellt uttryck. "Anledningen till att metoden är giltig är att gener som uttrycks vid låga nivåer eller med stor varians mellan replikat inte bidrar till den huvudsakliga mått som används av GSEA, den "anrikning värdering' (ES).
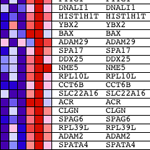
GSEA verkar genom att först rankning uttrycket värdet för varje gen genom Signal till brus ratio - beräkna skillnaden mellan medelvärdena för prov som representerar varje fenotyp och skalning dem med summan av standardavvikelserna. Detta innebär att gener med stora skillnader i uttryck nivå mellan olika stater och liten variation mellan biologiska replikat rankas högt.
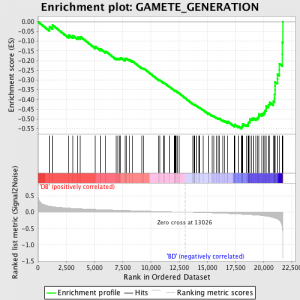
Nästa steg är att ES, den primära statistik som genereras av GSEA, beräknas för varje gen set - i GSEA manual, som dokumenterar programmet utmärkt, det står:
"Alla gener först rangordnade efter deras signal-brus-förhållande, då ES beräknas genom "walking" ner rangordnad lista av gener ökande en kör-sum statistik när en gen är i genen set och minskande den när den inte är. Den magnitud av ökningen beror på den korrelation av genen med en fenotyp. Det ES är den maximala avvikelsen från noll påträffas i walking listan. ETT positiv ES indikerar anrikning gen uppsättning på topp av rankningslista; en negativ ES indikerar anrikning gen uppsättning på botten av rankningslista. "
ES-värden är normaliserad baserad på genen som storlek och sedan en falsk upptäckt takt beräknas, för att ge en uppskattad sannolikhet för falska positiva. GSEA använder en mycket avslappnad standardvärdet 25%, som är lämplig för hypotes generation med ett relativt stort antal biologiska replikat.
Forskare som arbetar med uppgifter från non-human prover kan fortfarande använda GSEA, men behöverakta - Den genen symboler används av GSEA är "översatt"Från deras mänskliga motsvarigheter dvs. identifierare som används för gener från dina arter av intresse representerad på microarray omvandlas till symboler för deras humana ortologer, därefter användas i analysen. Subramanian och kollegor anspråk att denna omvandling har liten eller ingen effekt på användbarheten av GSEA; den har använts med framgång i flera icke-human art, men naturligtvis detta måste hållas i åtanke när man undersöker resultaten i detalj.
För en utmärkt, djupgående, översyn av pathway verktyg, samråda:
En annan bra källa för rådgivning på väg analys, särskilt för dem som är bekanta med R statistik paket är här.
Mer att läsa