 Gene Set Enrichment Analysis ist einer von vielen Ansätzen, um die Analyse der Genexpression Profildaten und ist unter a Papiervon Arbeitern am Broad Institute.
Gene Set Enrichment Analysis ist einer von vielen Ansätzen, um die Analyse der Genexpression Profildaten und ist unter a Papiervon Arbeitern am Broad Institute.
Das grundlegende Konzept wurde durch die Beobachtung, dass das Studium aufgefordert einzelne Gene zeigt die bedeutendste Unterschied in der Expression-Ebene zwischen zwei Staaten oder Phänotypen ist fehlt in mechanistische Einblicke. Stattdessen, macht es mehr Sinn, ein zu nehmen Satz von Genen sie einige biologische Verbindung, und die Frage stellen - nicht das ganze Set eine statistisch zeigen signifikante Anreicherung in jenen Genen, die differentielle Expression haben?
A Gen-Set gewählt werden kann, VON VORNHEREIN, für eine Anzahl von Gründen, z.B.. der Satz von Genen bekannt, um über beeinflussbar- oder Unterexpression eines Mikro-RNA, oder vielleicht ein Satz basierend auf chromosomale Standort gewählt, oder Gene für die molekulare Funktion, Zellkomponente und / oder biologischen Prozess zugewiesen wurden über die kontrollierten Vokabulare der Gene Ontology.
Ein Vorteil der GSEA Ansatz ist, dass es möglich ist, zu übernehmen Ihre vollständigen Datensatz, nicht nur diejenigen Transkripte mit einer willkürlich gewählten differentiellen Expression Schwelle. Ich bin mir sicher, dass viele Leute dies lesen werden denken - "Wie kann es sein OK, um den kompletten Datensatz zu verwenden? Normalerweise würde ich berücksichtigen nur Gene mit >2 (Oder andere Lieblings-Wert)-Falte unterschiedliche Expression. "Der Grund ist der Ansatz gültig ist, dass die Gene auf einem niedrigen Niveau oder mit einer großen Varianz zwischen repliziert ausgedrückt nicht von dem Haupt-Metrik durch GSEA verwendet beitragen, die 'Bereicherung des Gastes" (ES).
GSEA funktioniert durch die erste Rang der Wert des Ausdrucks für jedes Gen durch Signal-Rausch ratio - Berechnen der Differenz zwischen den Mittelwerten für die Proben, welche jedes Phänotyp und skaliert werden, damit durch die Summe der Standardabweichungen. Dies bedeutet, dass Gene, die mit großen Unterschiede in der Expression Ebene zwischen verschiedenen Staaten und wenig Variation zwischen biologischen repliziert werden hoch bewertet.
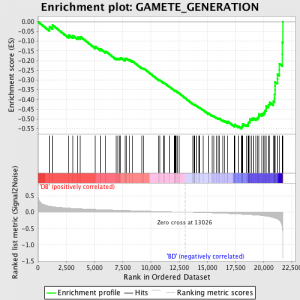
Der nächste Schritt ist, dass die ES, die primäre Statistik durch GSEA generiert, wird für jedes Gen Satz berechnet - im GSEA manuelle, Welche Dokumente die Software hervorragend, heißt es:
"Alle Gene werden zunächst durch ihr Signal-Rausch-Verhältnis sortiert, dann die ES durch "Fuß" down der Rangliste von Genen berechnet zunehmend ein Einlaufschicht Summe statistische wenn ein Gen in der Gen-Set ist und abnehmend es, wenn es nicht. Die Größenordnung der Inkrement abhängig von der Korrelation des Gens mit einem Phänotyp. Die ES ist die maximale Abweichung von Null zu Fuß die Liste gestoßen. A positiv ES gibt Gen-Set Bereicherung auf dem Top der Rangliste; ein negativ ES gibt Gen-Set Bereicherung auf dem Boden der Rangliste. "
Die ES-Werte sind normalisiert basierend auf Gen-Set-Größe und dann eine false discovery rate berechnet wird, um eine geschätzte Wahrscheinlichkeit von Fehlalarmen zu geben. GSEA nutzt eine sehr entspannte Standardwert 25%, welche geeignet ist Hypothesenerzeugung mit einer relativ großen Anzahl biologischer repliziert.
Wissenschaftler auf Daten aus nichtmenschlichen Proben können weiterhin GSEA, sondern müssensich hüten - Die Gensymbole verwendet von GSEA sind "übersetzt"Von ihren menschlichen Äquivalenten d.h.. Kennungen für Gene von eurer Spezies von Interesse auf dem Microarray vertreten eingesetzt werden zu Symbolen für ihre konvertierten Menschen Orthologe, dann in der Analyse verwendeten. Subramanian und Kollegen behaupten dass diese Umwandlung hat wenig oder kein Effekt über den Nutzen der GSEA; es wurde erfolgreich in mehreren nicht-menschlichen Spezies verwendet, aber natürlich muss im Auge behalten werden, in denen Ergebnisse im Detail.
Für eine exzellente, eingehend, Überprüfung der Weg Werkzeugen, konsultieren:
Eine weitere gute Quelle Erfahrungsberichte zu Pfadanalyse, vor allem für diejenigen, die mit der R Statistik-Paket ist hier.
Weiterführende Literatur