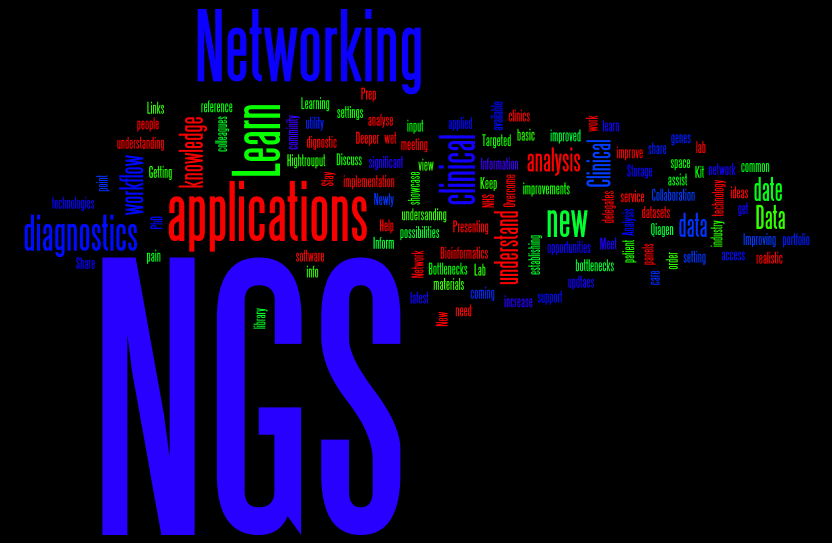
This word cloud shows the words that people used in the ice-breaking session, that I mentioned in my last post, to describe what they hoped to gain from the conference.
On Day 2, our Keynote speaker was:
Amongst other things, Dr Seller, Director of Genetics Laboratories, Oxford, told us how whole families benefited from better definition of the pathogenic potential of variants found using NGS, removing, in some cases, the need to continue clinical follow-up.
The next presentation, by Magnus Rattray, Professor of Computational & Systems Biology, University of Manchester, was a departure from the main theme of clinical genome sequencing and was entitled:
“Inferring transcript expression levels from RNA-Seq data and scoring differential expression between replicated conditions”
Prof. Rattray described how his group have used statistical methods to compensate for the “noise” and possible biases in data from the NGS of cDNA, known as RNA-Seq, in order to identify genes that are differentially expressed in various biological states. A link to one of the tools used, bitseq, is in the Tweet below:
These techniques are not so far from the clinic e.g. in some cancers, RNA-seq is being used to provide a signature that may discriminate sub-types. Returning us squarely into the clinic, however, was our next speaker, Dr. Klaus Brusgaard, Associate Professor Clinical Genetics, Odense University & Director at Amplexa Genetics, whose title was:
“Comparison of NGS Platforms & Software”
Dr Brusgaard guided the audience through the whole clinical NGS process, from sample handling to final analysis. Many of the illustrative examples used were from patients with epilepsy, especially childhood onset, where targeted sequencing of a gene panel was shown to be very powerful.
Our next speaker, Mick Watson, from ARK-Genomics at the University of Edinburgh, reminded us that humans are not only primates, but also hosts to a microbial community – our guts contain approximately ten times more bacteria than the number of eukaryotic cells in the rest of our bodies!
However, as you can tell from the shortened title in the Tweet above, Mick’s topic was the NGS and analysis of the combined genomes of the microbes (metagenome) found in the guts of ruminants. He showed some compelling data that implied that we don’t need to see a reindeer to differentiate it from, say, a dairy cow, instead we could tell them apart by analysis of the metagenome sequences in their guts. Mick also briefly discussed the manifold influences of gut microbes on their hosts.
Les Mara, from Databiology, gave the next presentation, entitled:
“Avoiding the ‘Omics Tsunami”
- a cautionary tale about the importance of robust, scalable and easily deployed Bioinformatics solutions, in order to not only withstand the Big Data deluge, but to extract best value from your investment.
The last talk before lunch on Day 2 was given by Kim Brugger, Head of Bioinformatics, EASIH, University of Cambridge, who had changed the title of his talk from:
to:
“Managing IT resources and NGS LIMS for free”
The emphasis of this talk was how free, open-source software could be assembled into a potent pipeline handling a variety of NGS data, tracking all updates, upgrades or other changes that would require re-validation and still confer confidence in results from one run to another.
The afternoon session was kicked off by Dr Jim White, NanoString Technologies, who took us beyond NGS, to a technology aimed at following-up on copy-number variants or differences in expression discovered in the research or clinical NGS lab:
TECHNOLOGY WORKSHOP – “NanoString hypothesis driven research tool drives NGS Discoveries towards the Clinic”
Using fluorescent probe hybridisation-capture, up to 800 different target RNA transcripts or genomic sequences can be quantified.
Our next speaker, Kevin Blighe, Lead Bioinformatician at the Sheffield Children’s NHS Foundation Trust, told us about:
He gave us a warts-and-all point of view of introducing NGS in a clinical environment, including the challenges of the perceptions of the UK National Health Service reported by the media…
The penultimate presentation of the day came from – me! I gave the audience my point of view on the difficulties inherent in:
“Predicting Effects of Sequence Variants on Phenotype”
Inspired by a blog post, I gave an overview of the challenges and then attempted to answer some of these, emphasizing the importance of model organism studies to improve understanding of genetic background effects, gene-environment interactions and non-coding variants.
Our last talk was given by:
Post-Doctoral Research Fellow in the School of Biomedical Sciences, University of Leeds. Dr Ivorra-Martinez told us how a carefully constructed family pedigree and a good understanding of the inheritance pattern of schizophrenia, when combined with exome sequencing allows the identification of novel, likely causative, variants.
Based on the feedback that we received, the conference and workshops achieved many of our aims, in particular, stimulating conversations and offering opportunities for partnerships – perhaps we will see you next time?!
 I recently returned, albeit part-time, to work in academia and I was offered the chance to teach. Many academics in scientific research never teach, other than perhaps one-to-one “teaching” of a new doctoral student. In the past, I have presented my work to other scientists and also given a few guest lectures to undergraduates; but this was the first occasion when the value and importance of teaching really hit me and I’ve tried to capture that below. So, why do I think science researchers should teach?
I recently returned, albeit part-time, to work in academia and I was offered the chance to teach. Many academics in scientific research never teach, other than perhaps one-to-one “teaching” of a new doctoral student. In the past, I have presented my work to other scientists and also given a few guest lectures to undergraduates; but this was the first occasion when the value and importance of teaching really hit me and I’ve tried to capture that below. So, why do I think science researchers should teach?